User documentation
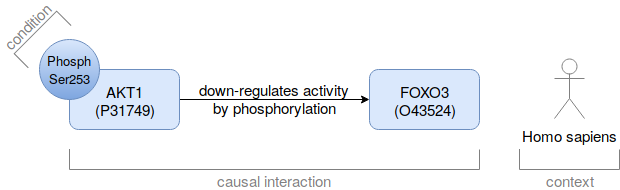
What is a molecular interaction causal statement?
In simple words: It is an interaction between biological entities where a source entity regulates the activity of a target entity in a specific context.
In more detail: A molecular interaction causal statement can be decomposed into a molecular interaction and a causal statement. The molecular interaction covers undirected (i.e., non-oriented) interactions between biological entities (e.g., protein, gene, complex), and does not have to be necessarily physical interactions. The causal statement adds a causal aspect to the molecular interaction with a regulatory aspect and a direction of the regulation, i.e., a biological entity A (the source) interacts with and has an effect on a biological entity B (the target). Furthermore, a causal statement describes contextual details that inform on specific circumstances in which the causal effect is observed.
To know more about the minimum set of information to annotate about a molecular interaction causal statement, read the MI2CAST guidelines.
What is VSM (Visual Syntax Method)?
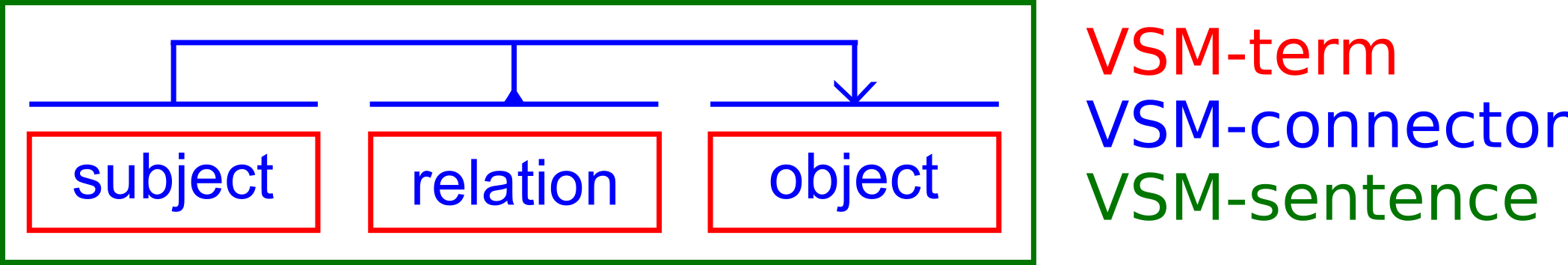
VSM is a general framework for communicating pieces of information that are both human and computer understandable. VSM includes three component types:
1- a VSM-term, which is an association of a term (human-readable) and an identifier (machine-readable),
2- a VSM-connector, which is a grouping of VSM-terms based on a particular semantics (for example subject-relation-object),
3- a VSM-sentence, which consists of an association of VSM-terms connected with VSM-connectors.
The causalBuilder is built on top of the VSM framework to facilitate the annotation of causal statements with contextual information. The causalBuilder generates on-demand templates of VSM-sentences to be filled in by the user.
To know more about VSM, see the VSM overview.
How to use the web application?
The causalBuilder is divided into three parts:
1- the selection of contextual annotations to add in the causal statement,
2- the template needs to be filled in manually by the user with the adequate data,
3- download the annotated causal statement.
Go to the causalBuilder home page and follow the steps below:
1. Select all the annotations (context) to add in the causal statement
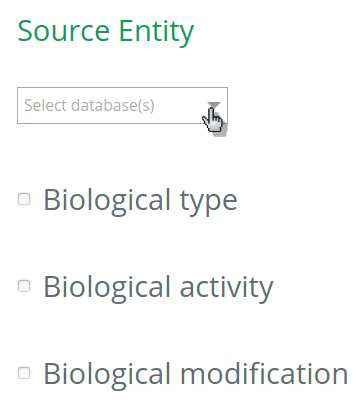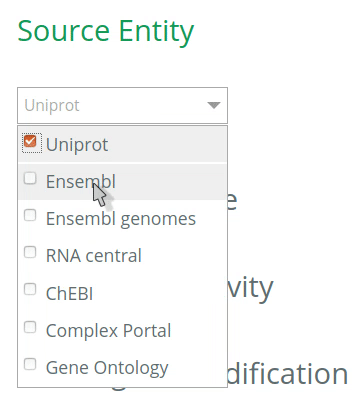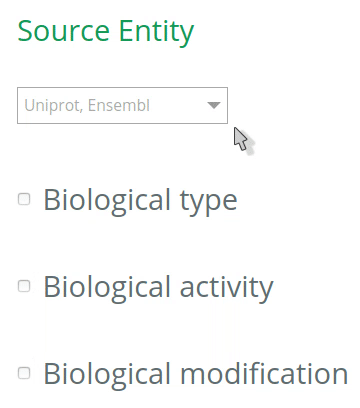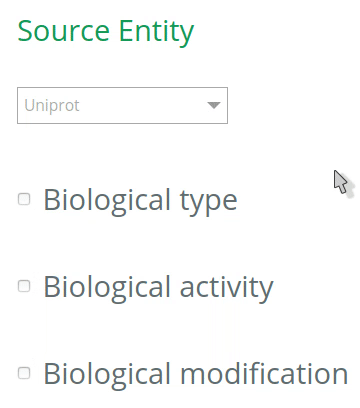
1.1 Select the database to annotate your entities
By default, all databases will be checked for the annotation of entities (UniProt, Ensembl, Complex Portal, RNAcentral, etc). To restrict the options (and get more accurate terms in the auto-complete when filling the VSM-template at step 2), click on the desired database(s) for the annotation of your source and target entities. The selection should be based on the biological type of the entity. For instance, if your source entity is a protein, check the UniProt under Source entity, then the options in the VSM-term of the ‘source’ will be restricted to UniProt identifiers only (step 2).
1.2 Adding a term from a checkbox
Click on the desired checkbox to select annotation to be added in the causal statement. For instance, if you would like to annotate the biological type of a source entity, select the biological type checkbox under Source Entity.
1.3 Adding a biological modification
A biological entity, whether it is a source or target entity can have multiple biological modifications annotated. The biological modifications correspond to physical modifications or conformations of an entity (e.g., phosphorylation of a protein, methylation of a gene, etc.). To add a biological modification, whether it belongs to the source entity or the target entity, first, check the biological state box and then select the type of annotation in the drop-down list that appears:
1.3.1 mod
Biological modification with information on the type of modification only. Modification corresponds to the chemical modification affecting the entity (e.g., phosphorylation, acetylation).
1.3.2 mod+res
Biological modification with information on the type of modification and the residue affected. The residue corresponds to the modified amino acid of the entity (e.g., histidine, serine).
1.3.3 mod+pos
Biological modification with information on the type of modification and the position. The position corresponds to the position of the modified amino acid in the entity.
1.3.4 mod+res+pos
Biological modification with information on the type of modification, the residue and, the position.

Once the biological modification is checked, a new one appears below to enable the annotation of another biological modification. If the checkbox is not checked and if the type of annotation (i.e., mod, mod+red, mod+pos, mod+pos+res) is not selected, the terms will not appear in the VSM-sentence.
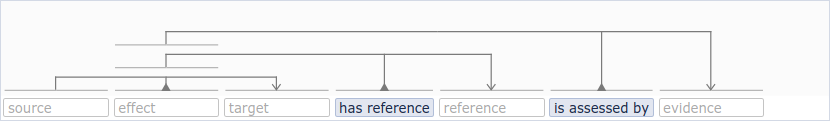
1.4 Adding experimental setups(s), reference(s) and evidence
A causal statement has at least 1 references and/or evidence, but can also have more. A source entity and a target entity can have 0, 1 or many experimental setups defined, which correspond to particular experimental setting(s) applied to the entities to observe the causal interaction. Consequently, indicate the number of reference, experimental setup and evidence metadata wished to annotate by entering a number or by using the arrow on the right side of the box. By default, the amount is set to zero for experimental setup and to 1 for reference and evidence.

The VSM-sentence “template” is progressively built as the annotations are selected. When a checkbox is checked, a type selected or a number added in the reference/evidence/experimental setup, you can see that the vsm-box is automatically updated to insert the blocks of information to be annotated.
2. Fill the VSM-template
Click on a VSM-term and start typing the identifier or the name of the annotation. When the corresponding match appears, select the appropriate annotation term and click on it or press ENTER. Fill the other annotations accordingly.
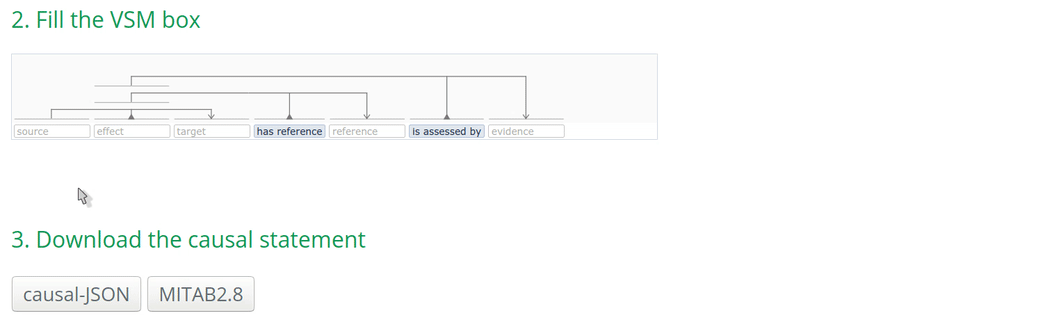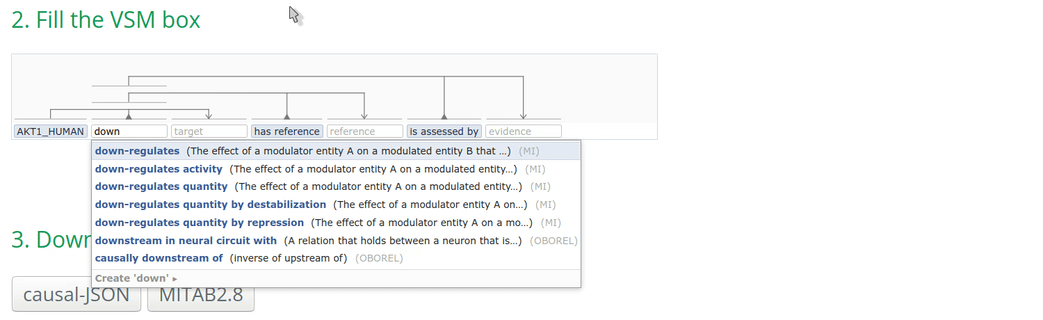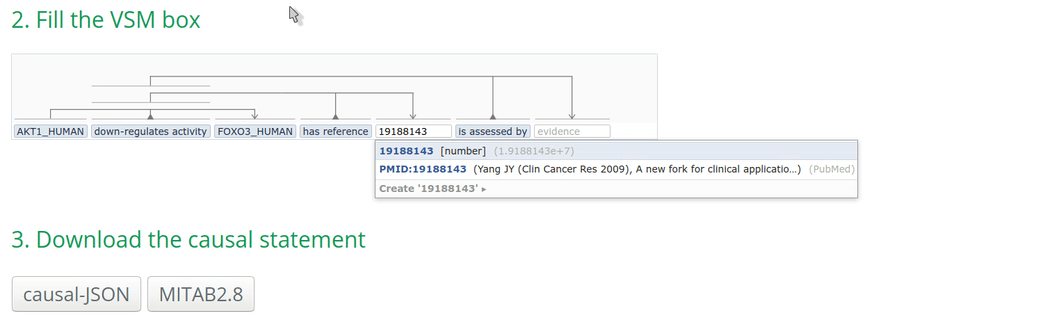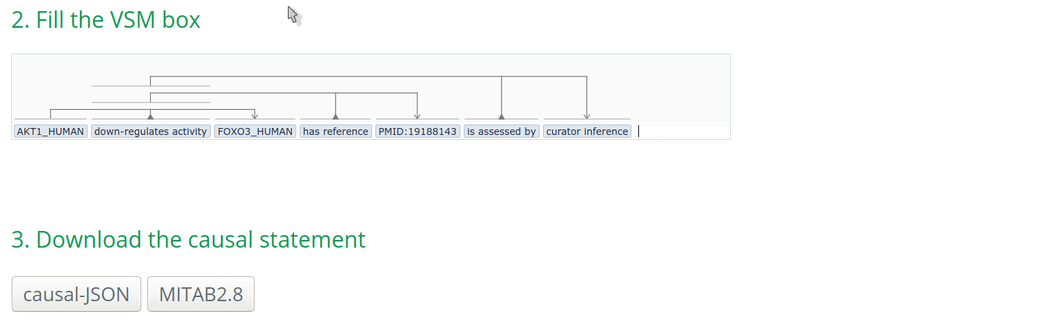
Note: Double-clicking on an empty VSM-term field shows the vocabularies or ID lists that the auto-complete for that specific field is configured for. The list of possible annotations for a term can also be found at https://github.com/MI2CAST/MI2CAST.
3. Download the causal statement in the desired format
The causalBuilder can generate causal-JSON and a MITAB2.8 formatted files. In the causal-JSON, all information that a curator filled in the VSM-template will be translated and included in full. In the MITAB2.8, only the information that can be supported by this format is stored (for instance, the experimental setup can currently not be stored). Check the MITAB2.8 documentation for more information.
In addition, causalBuilder can export the VSM-JSON format, which is a direct representation of the VSM-sentence or -template data emitted by the vsm-box. The VSM-JSON-‘light’ format is a subset of core data (including IDs and term names but excluding some template-related data like autocomplete filters), and constitutes a concisely formatted and human-readable essence of what the curator has annotated.
Questions? Problems? Contact us!
Do not hesitate to reach us via the GitHub platform.