Some tips to effectively annotate a molecular causal interaction’s different aspects, using causalBuilder.
How to annotate an entity?
- Select the database based on the type of entity to be annotated. For instance, to annotate a protein, select UniProt.
- There are several ways to optimize the search for a specific entity in the VSM-box, from the most precise to the less specific search:
- search by the identifier (e.g., P31750) which should always give you only one result
- search by the name (e.g., AKT1) which may result in several options.
Tip: for UniProt, using the ‘Entry name’ provides always only one result! For instance, AKT1_mouse:
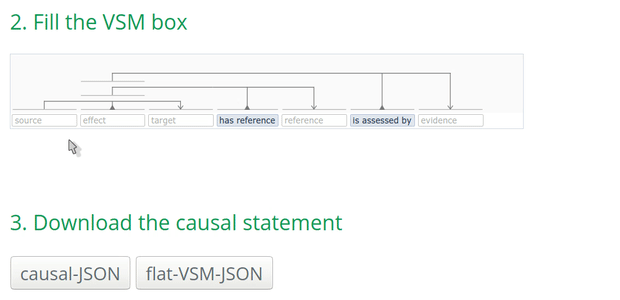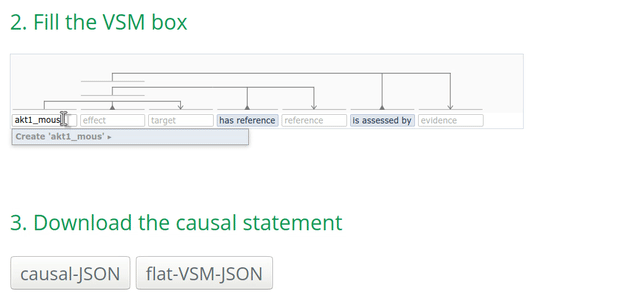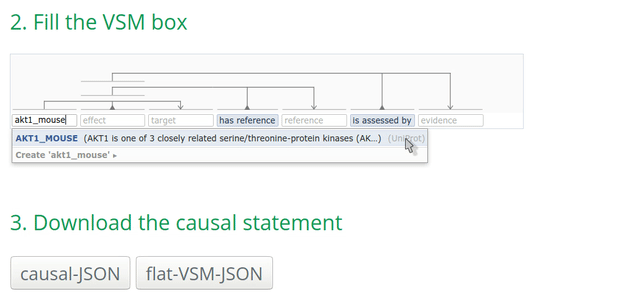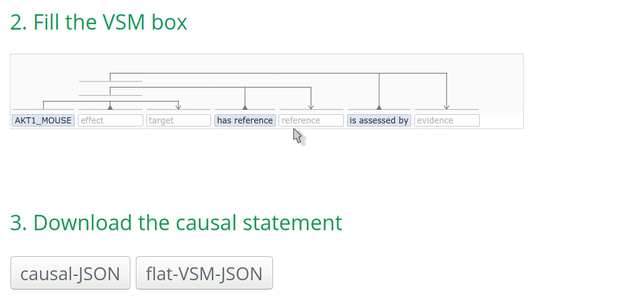
How to annotate the biological type of an entity?
By default, the biological type of an entity is determined by the ontology used. For instance, a source entity annotated with a UniProt identifier is considered as a protein. The mapping of ontologies and biological type is provided in Table 1 below:
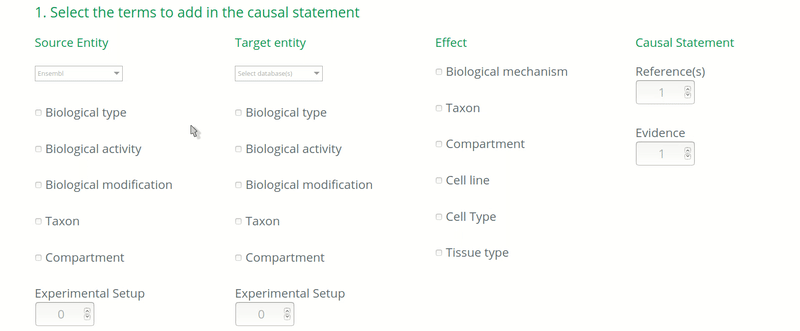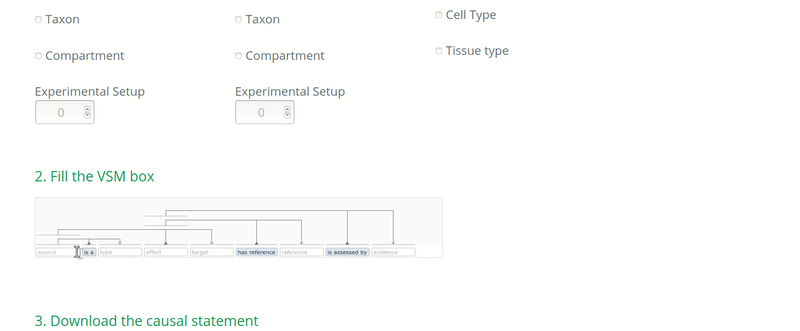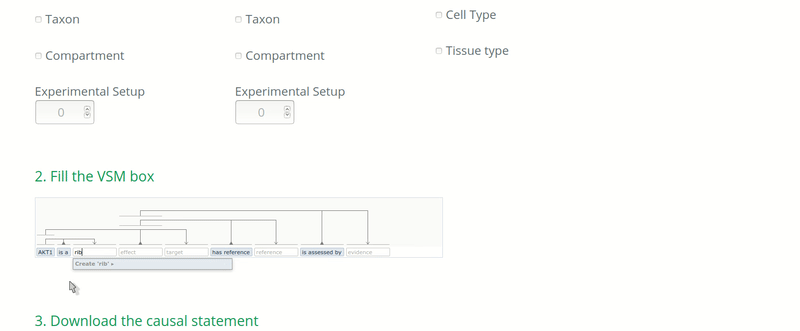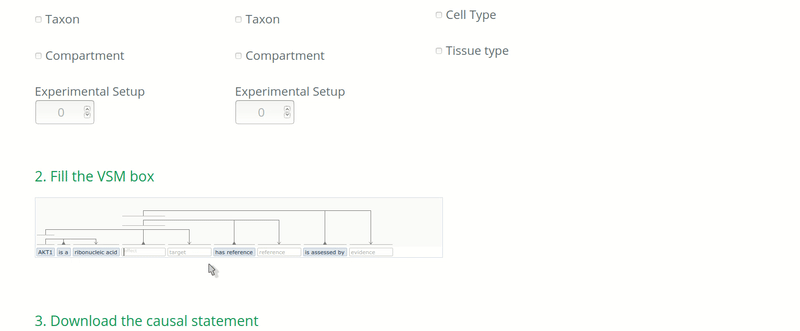
In the case when the curators know that the ontology used to annotate an entity DOES NOT correspond to its real biological type, then the real biological type of the entity should be annotated. For instance, if an article assesses the causal regulation of a transcript but the transcript identifier is unknown or not mentioned in the paper, then the curator would use the identifier of the gene to annotate the target entity, because the gene is most likely to be known. In this case, the biological type of the target entity should be annotated as “ribonucleic acid” (http://purl.obolibrary.org/obo/MI_2233) if following the PSI-MI.
How to annotate the taxon?
By default, the taxon of the entities corresponds to the ones determined by the identifier given for the entity. For instance, if the source is annotated as ‘P06493’, corresponding to CDK1_HUMAN, then the source is considered to come from a human. If the taxon from the annotated identifier does not correspond to the observed one in the causal statement, then it must be specified. In this case, the taxon of the entity should be annotated with an NCBI Taxonomy identifier of the corrected taxon.